Multi-scale simulation and modeling of biomolecular processes
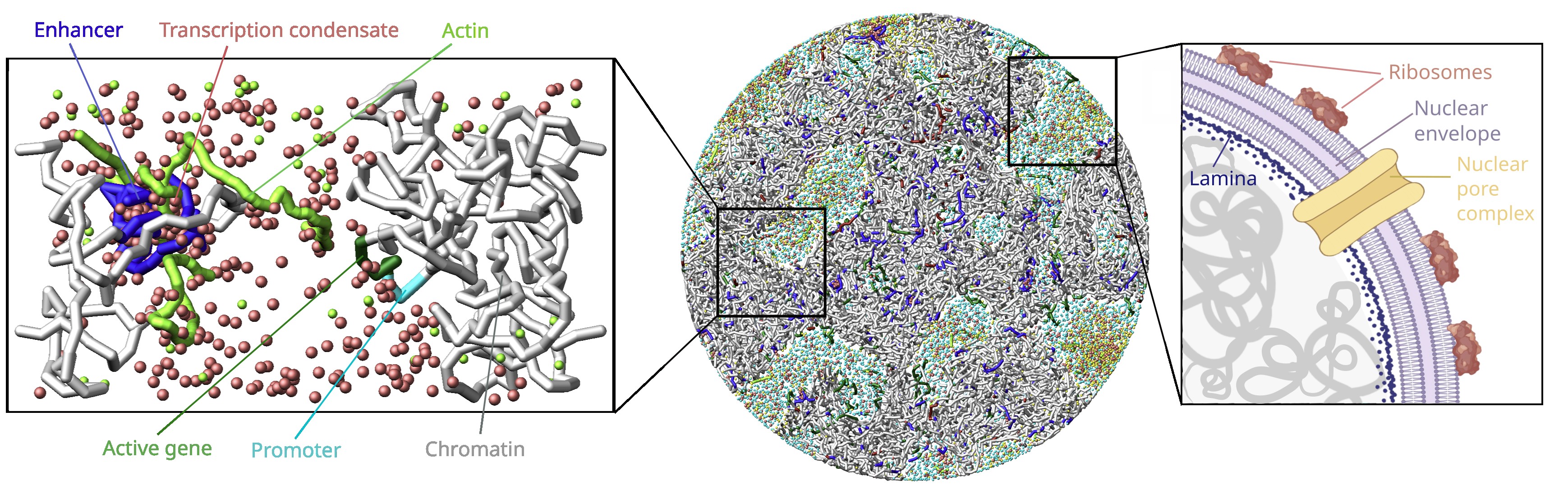
The cell nucleus is a densely packed, dynamic and spatially structured environment where molecular interactions, mechanical forces, and phase-separated compartments jointly control genome function. Understanding these highly dynamic processes requires approaches that systematically explore mechanisms beyond the reach of laboratory experiments. Computational modeling and molecular simulations provide exactly this capability, offering quantitative, mechanistic insights into nuclear organization and gene regulation by linking specified interaction rules to biomolecular behavior at the cellular scale.
Our group develops computational models and simulations that reveal the physical principles underlying nuclear organization and its impact on gene expression. By integrating coarse-grained molecular dynamics, multiscale biophysical modeling, and validation experimental data, we aim to construct predictive frameworks that bridge molecular interactions to whole-nucleus function.
Our interdisciplinary work is deeply integrated with the departments of IBCS, especially the advanced microscopy imaging in the Department “Information”, where research on stem cells and zebrafish embryos provides key dynamical and structural data for model validation. Additional experimental cooperation expand our ability to test and refine synthetic and biomimetic systems inspired by nuclear processes.
Simulation Framework of the Cell Nucleus
We develop molecular dynamics–based simulation framework of nuclear architecture and intranuclear processes using LAMMPS embedded in Python. Our model currently integrates transcription regulation through transcription condensates, actin-guided gene recruitment, and spatiotemporal switching between regulatory states, with ongoing research focusing on elucidating the morphology of transcription clusters as well as the details of actin role in transcription regulation. Parametrization and validation are carried out in close alignment with experimental data from zebrafish embryos to ensure biological realism.
Actin-Guided DNA Transport and Synthetic Genome-Inspired Systems
In collaboration with experimental partners at KIT and Johannes Gutenberg University Mainz, we are aiming to develop simulations to explore how phase-separating DNA nanostars can be coupled to engineered actin networks, creating a synthetic analogue of long-range enhancer–promoter communication in early embryos. By combining coarse-grained modeling with experimental reconstruction, we want to investigate how network architecture, DNA sequence encoded droplet properties, and actin-driven mechanics control transport efficiency, cargo selection and delivery specificity. This research supports a broader vision of building programmable, genome-inspired soft-matter systems capable of long range, sequence-selective transport.
Modeling the Nuclear Envelope as an Active Regulatory Interface
We are currently extending our whole nucleus simulation platform to include an explicit and mechanistically detailed representation of the nuclear envelope and nuclear pore complexes (NPCs), aiming to redefine its role from a passive boundary to a dynamic, active mechanochemical interface. This multiscale modeling approach combines coarse-grained molecular level simulation with targeted atomistic parametrization to address the following research topics:
- The role of lamina organization in regulating chromatin tethering and genome architecture
- Mechanotransduction across cell nucleus and nuclear membrane, and its impact on chromatin transcriptional accessibility
- The impact of NPC mechanics and structure on selective RNA-protein transport
Methods and Tools
- Coarse-grained molecular dynamics and modeling (LAMMPS, Python)
- oxDNA for higher resolution DNA and polymer simulations
- Close collaboration with wet-lab groups for model refinement and hypothesis testing
Join Us
We welcome Bachelor and Master students and researchers with interests in:
- computational biology and biophysics
- molecular modeling and simulation
- genome organization and transcription dynamics
- multiscale modeling of complex biological systems
- theory - experiment integration
Whether your background is in physics, computational biology, applied mathematics or a related field, we offer a collaborative space for interdisciplinary science.
Ewa Anna Oprzeska-Zingrebe, PhD
Karlsruhe Institute of Technology (KIT)
Institute of Biological and Chemical Systems (IBCS-FMS)
Zoological Institute (ZOO)
Hermann-von-Helmholtz-Platz 1
Building 319
76344 Eggenstein-Leopoldshafen, Germany
E-Mail: ewa.zingrebe∂kit.edu

